Exploring GIs in a Human Cell Line:
Mapping a Genetic Network for a Reference Human Cell Line
This project is a collaboration with the lab of Jason Moffat, Genetics and Genome Biology at Sick Kids
People
Michael Costanzo, Ermira Shuteriqi, Wendy Liang, Ryan Climie, Lyudmila Nedyalkova, Matej Usaj
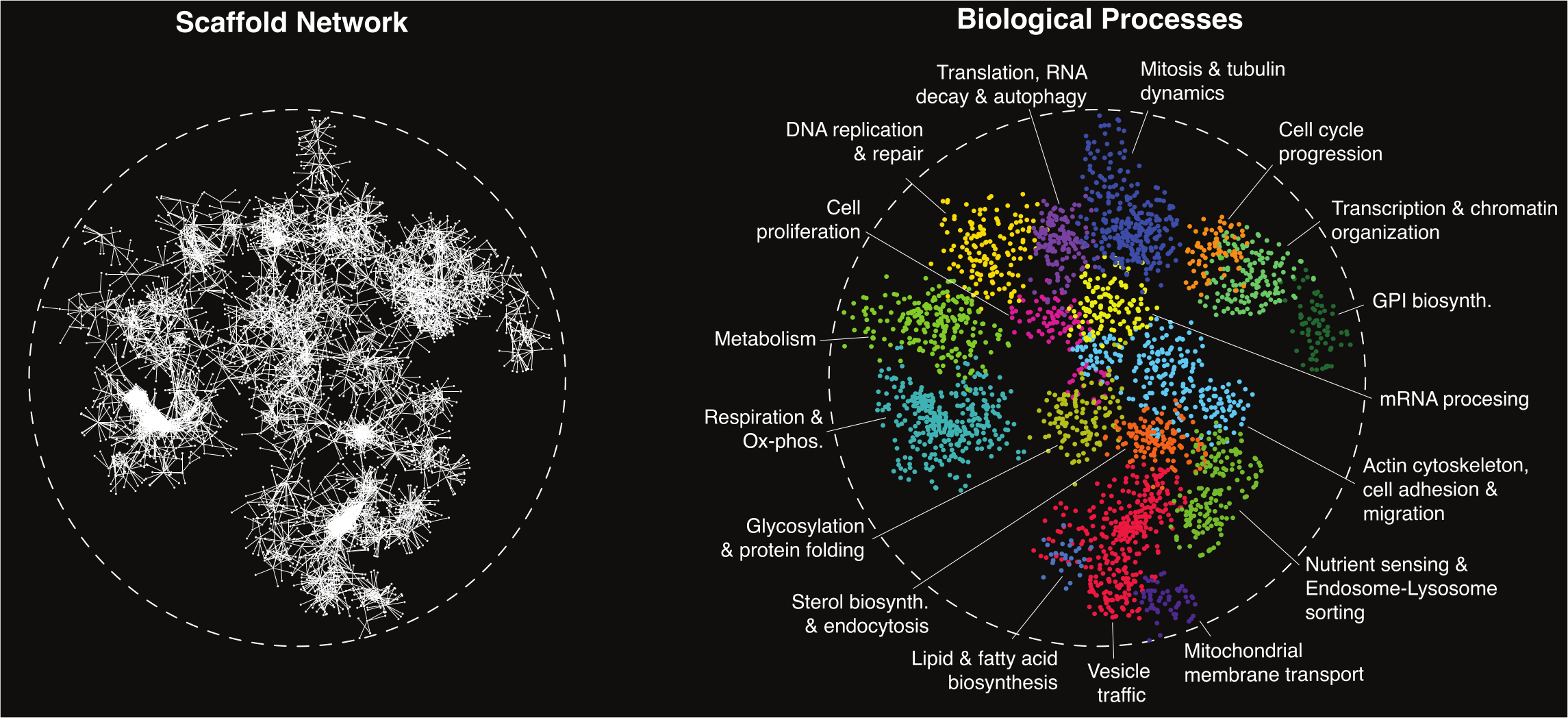
Genetic interactions identify functional connections between genes and impact the relationship between genotype and phenotype. Previous application of an automated genetic approach, called Synthetic Genetic Array (SGA) (Tong, 2001, 2004), identified ~1 million quantitative negative and positive genetic interactions among the majority of all possible gene pairs in the budding yeast model eukaryote, Saccharomyces cerevisiae (Costanzo, 2010, 2016). Negative interactions, such as synthetic lethal or sick interactions, occur when a double mutant shows a fitness defect greater than the expected effect for the combined single mutant fitness phenotypes and positive interactions reflect double mutants that grow better than expected (Costanzo, 2019). A global network based on profiles of negative and positive genetic interactions grouped genes into sets of hierarchically organized modules corresponding to protein complexes or pathways, biological processes and cellular compartments, revealing the functional architecture of a yeast cell. Moreover, while only ~1000 (~20%) genes are individually essential for yeast cell viability, systematic genetic interaction analysis identified ~10-fold more extreme digenic synthetic lethal interactions, where a nonessential gene becomes indispensable for growth in the context of a specific genetic background, emphasizing the immense potential for genetic interactions to modulate phenotype (Costanzo, 2010, 2016).
Development of CRISPR/Cas9 gene-editing technologies enables analogous large-scale genetic interaction studies in human cells. We are using CRISPR gene editing methods to systematically introduce a second mutation into a collection of engineered mutant cell lines, each carrying a stable mutation in a single "query" gene of interest, to map genome-scale genetic interaction network in a human HAP1 cell line. Genetic interactions tend to connect functionally related genes and members of the same pathway or complex often share similar patterns of genetic interactions. Indeed, genes with correlated genetic interaction profiles formed discernible network clusters that are enriched for specific cellular functions suggesting that a human genetic network, like the global yeast network is rich in functional information, and provides a powerful approach for annotating human gene function.