Building and Characterizing New Yeast Collections:
Sequence Analysis of our Temperature-sensitive Mutant Collection and Construction of a GFP-tagged Version of the TS Collection
People
Zhijian Li, Dale Climie, Hong Lu, Kyle Wang and Myra Paz Masinas
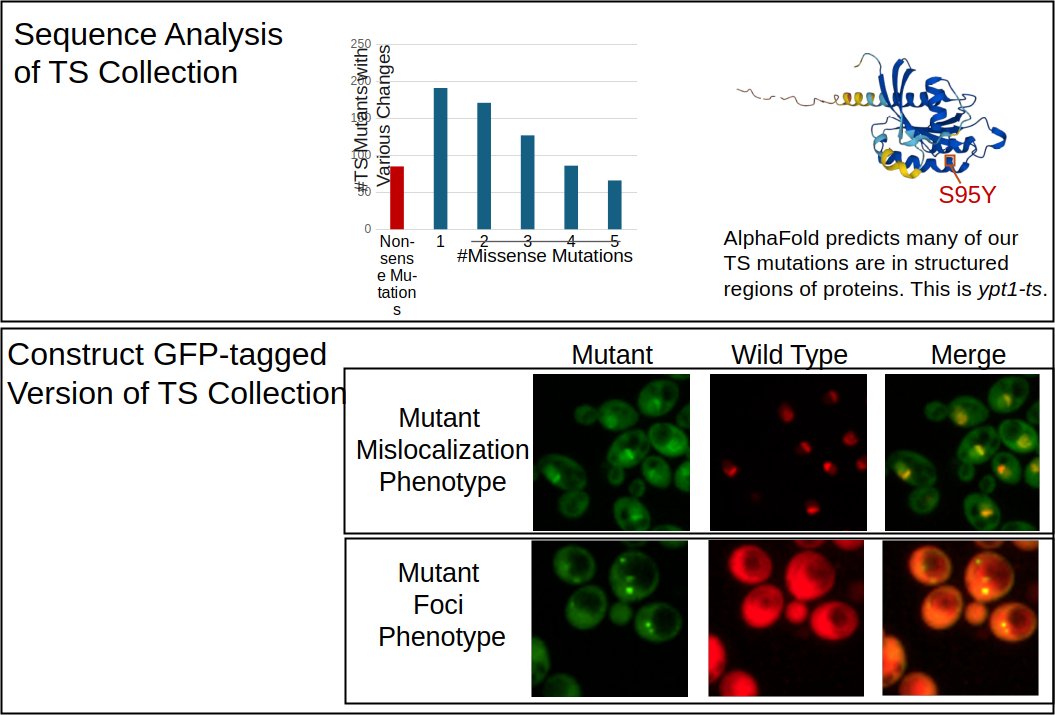
We have previously constructed temperature-sensitive (TS) mutants for all ~1000 essential genes in yeast. Most of these TS alleles are hypomorphic, even at permissive temperature. Now, to understand what amino acid changes confer these mutant phenotypes, we have sequenced the TS collection and are using structure prediction and variant effect mapping tools to explore the data. In addition, to identify the consequences of each mutation on the localization and abundance of the protein in vivo, we have constructed GFP fusions of all the TS alleles.
Despite cataloging millions of missense variants across human genomes, how these mutations affect phenotypes at the protein level is not well understood. Abnormal protein phenotypes such as subcellular mislocalization, changes in abundance and aggregation have been hallmarks of many diseases, rare and common, that affect human health today. To gain an understanding of this genotype to phenotype relationship, we need to examine a broad spectrum of abnormal proteins and catalogue their phenotypic landscape. To do this, we have constructed a library of over 1000 essential gene mutants, where each mutant allele was fused to a fluorescent protein. We imaged every mutant with its wild type counterpart in 12 hr time course experiments, capturing dynamic protein localization and abundance data in millions of live cells. We trained a convolutional neural network to classify subcellular localization and extracted protein abundance data at the single cell level. Using this dynamic dataset, we aim to systematically explore the phenotypic landscape of abnormal proteins in the cell and decipher the rules that govern how mutations at the DNA level results in subcellular phenotypes.