Building and Characterizing New Yeast Collections:
Constructing Deletion Collections in Natural Yeast Isolates Using CRISPR-Cas9
People
Guihong Tan, Fang He
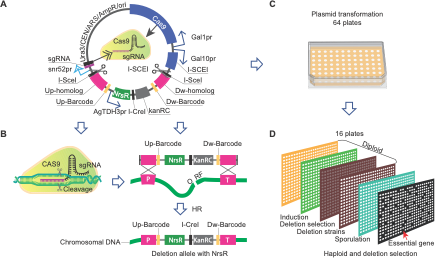
Different wild yeast strains are specialized for different niches, which is reflected by their different genetics. To gain comparative insights into background-dependent fitness phenotypes and associated modifier interactions, and to dissect the core essential genes of yeast requires the generation of systematic deletion mutant collections in genetically diverse strains. Traditional PCR-based is costly and can be a major bottleneck at this scale. We have developed a collection of CRISPR-Cas9 based gene deletion plasmids consisting of ~5800 individual plasmids (> 98% of the genome), each of which targets and deletes a unique gene in the yeast genome through homology-based repair. These plasmids are designed to take into account the sequence divergence among different wild strain backgrounds, such that the guide RNA can target every strain in our collection despite their genome diversity, and the repair fragments include large homology regions of ~200 bp ensuring efficient repair in any background. We can score gene essentiality across natural yeast isolates individually in an array format. In addition, deletion mutants generated with these plasmids carry unique 20 bp DNA barcodes, which allows for their identification using a pooled barcode sequencing strategy. With these resources, we are focusing on ~9 diverse genetic backgrounds to generate systematic gene deletion collections.